JBRA Assist. Reprod. 2015; 19 (2):37-43
ORIGINAL ARTICLE
doi: 10.5935/1518-0557.20150011
Triplex Real-time Polymerase Chain Reaction Optimization for AZF Y-chromosome Microdeletion Analysis
1Centro Nacional de Reproducción Asistida INNAIFEST – Guayaquil - Ecuador
2Laboratorio de Biología Molecular de la Facultad de Medicina. Universidad Espíritu Santo - Ecuador
CONFLICT OF INTERESTS
No conflict of interest have been declared.
ABSTRACT
Objective: Y chromosome microdeletions at the “Azoospermia Factor” regions (AZFa, AZFb, AZFc) are the second genetic cause of spermatogenic failure in infertile men. Despite its importance for the treatment of infertile patients, no prior investigations have been previously published in Ecuador. . The purpose of this study is to optimize a molecular technique that allows detection of microdeletions in the AZF region.
Methods: Using a genomic DNA of healthy male with natural conceived offsprings, a multiplex real time polymerase chain reaction (qPCR) was standarized with eigth sequence-tagged site (STS) sY85, G34990, sY133, sY127, sY254, sY255, and using as internal control sex-determine region Y (SRY) and Ameologenin Y (AMELY). With this technique, 35 DNA samples taken from peripheral blood of patients with severe oligozoospermia were analyzed.
Results: A triplex qPCR was standardized using EvaGreen DNA-binding dye to obtain melting temperature (Tm) of the STS previously mentioned. Three of the patients evaluated were detected to have partial microdeletion in the AZFa region, with a frequency of 8.8%; being losses in the G34990 section (one patient) and sY85 section (two patients). No cases of microdeletions in other AZF regions were found.
Conclusion: The triplex qPCR optimizated allows the identification of microdeletions in AZFa, AZFb and AZFc region in infertile men and a better clinical management of the patient’s treatment decision. This first report for Ecuador reveled a higher prevalence of microdeletions in the AZFa region in comparison with those previously described in other populations.
Keywords: AZF, Microdeletion, PCR, Y Chromosome, Male infertility.
INTRODUCTION
Infertility affects about 15% of couples worldwide, 50% of these cases due to male factors (Elfateh et al., 2014; Kovac et al., 2013; Sheikhha et al., 2013; O’Flynn et al., 2010; Kozina et al., 2011; Dohle et al., 2010).
Within the etiology of male infertility, the genetic factor plays an important role, influencing in a variety of physiological processes such as spermatogenesis and sperm quality. This factor would represent between 15 to 30% of the causes of male infertility (Elfateh et al., 2014; Khabour et al., 2014; O’Flynn et al., 2010; Dohle et al., 2010; Kleiman et al., 2012), being microdeletions of the Y chromosome (YCMD) the second genetic cause of spermatogenic failure in infertile men, after Klinefelter syndrome (Khabour et al., 2014; Kozina et al., 2011; Guo et al., 2012; Silber, 2011).
In the Y chromosome, there are three regions referred to as “Azoospermia Factor” (AZFa, AZFb, AZFc), all related to spermatogenesis and normally altered or lost in YCMD, this is found with a frequency of 13% in men with non-obstructive azoospermia and 7-10% in men with oligozoospermia (Yuen et al., 2014; Zaimy et al., 2013; Kozina et al., 2011;. Guo et al., 2012; O’Flynn et al., 2010; Soares et al., 2012).
The AZFa region contains two major genes involved in fertility, these are USPY and DBY. The total deletion of this region is rare (5%) and only a few patients have been described. It is associated with the complete absence of germ cells, presenting Sertoli cells-only syndrome (SCO) (Sheikhha et al., 2013; Krausz et al., 2014; Suganthi et al., 2013; Silber, 2011; Kleiman et al, 2012; O’Flynn et al., 2010). In these cases it is impossible to retrieve sperm from the testes for assisted reproduction treatments (ART) such as intracytoplasmic sperm injection (ICSI) (Guo et al., 2012).
Partial deletions of AZFa have a wider range of prevalence (0.2%-11%) and its histological characteristics range from normozoospermia to SCO (Kleiman et al., 2012), depending on the clinical syndrome, the man may or may have not have problems in producing offspring.
In the AZFb region several genes are found, being the main RBMY and PRY. Deletions in this area are more common (10%-16%) than those of the AZFa region, nevertheless these are found in a very low percentage of azoospermic men (Krausz et al., 2014; Suganthi et al., 2014; Silber, 2011; Soares et al., 2012). The total deletion of the region completely stops spermatogenesis, causing azoospermia and total absence of sperm in the testicles. This would imply not have sperm through testicular sperm extraction (TESE) process (O’Flynn et al., 2010; Silber, 2011).
Rolf et al. (2002) have reported the presence of mature sperm cells with deletions in the region AZFb, associating microdeletions in such area with moderate oligozoospermia, allowing natural conception and therefore the transmission of this genetic abnormality. It is possible to find sperm cells in men with partial deletions in this region (Krausz et al., 2014; Suganthi et al., 2013; Kleiman et al., 2011).
Deletions in AZFc region are the most frequent (50% and 60%) (Kleiman et al., 2011; Soares et al., 2012; Silber, 2011). They produce a wide range of phenotypes associated with low sperm concentration and reduced spermatogenesis, being found both in azoospermic or severely oligozoospermic men (O’Flynn et al., 2010; Guo et al., 2012; Navarro-Costa et al., 2007). This suggests that genes located in this area (mainly the DAZ family) are involved in the maturation of post meiotic germ cells or sperm (Soares et al., 2012; Silber, 2011).
For men who are azoospermic with deletions in the AZFc area, it is possible to obtain sperm through TESE and to conceive by ICSI, considering that Y chromosome genetic disorders would be transmitted to the male offspring (Kleiman et al., 2011; Guo et al., 2012).
The diagnosis of any YCMD can clarify the cause of idiopathic non-obstructive azoospermia or oligozoospermia, influencing over the treatment options (Kozina et al., 2011; Dohle et al., 2010; Guo et al., 2012).
Current methods for diagnostic are based on polymerase chain reaction (PCR), amplifying specific sequence sites (STS) within the region of interest. The European Academy of Andrology (EAA) and European Network of Molecular Genetics Quality (EMQN) published the guide for the molecular diagnosis of Y chromosome microdeletions (Krausz et al., 2014; Kozina et al., 2011; Guo et al., 2012; Simoni, 2004, 2001).
Despite the importance of the evaluation of this type of microdeletions in infertile men, no prior investigation has been published in Ecuador about this topic.
The aim of this estudy is to optimize a molecular technique that allows detection of microdeletions in the AZF region.
MATERIAL AND METHODS
Patients
Peripheral blood samples from 35 oligozoospermic men without offspring were obtained from the National Center of Assisted Reproduction Innaifest. For Positive control, peripheral blood samples from two fertile men (25 and 23 years old) with naturally conceived children, were considered. The semen parameters were evaluated acording WHO (2010).
Isolation of DNA
Genomic DNA were extracted using High Pure PCR Template Preparation Kit, version 16.0 (Roche: Cat. No. 11 796 828 001) according to the manufacturer’s protocol. The concentration of extracted DNA was determined by measuring the UV absorbance at 260 nm with the “Biophotometer” spectrophotometer (Eppendorf).
Primers
Eight primer pairs were setected for the publication of Kozina et al. (2011) (Figure 1). Two pairs were used as internal control and six primer pairs were specific to the sites AZFa, AZFb and AZFc (Table 1).
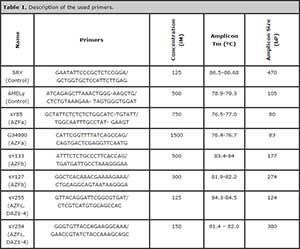
Table 1. Description of the used primers.
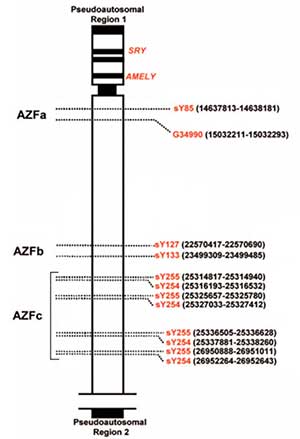
Figure 1. Location of the Primers used for the multiplex PCR, in the Y chromosome. Modified scheme from Kozina et al. (2011). On the left side the AZFa, AZFb, AZFc regions can be identified. On the right the union sites for the primers are identified in red and their genomic position is shown in black.
Multiplex Real-Time PCR
The study was carried out in three triplex real-time PCR using the Mastercycler, Version 2 Channels 520 nm / 550 nm (Eppendorf). PCR conditions were 95ºC for 60 seconds, followed by 35 cycles of 95ºC for 15 seconds, 60ºC for 45 seconds, 72ºC for 30 seconds and a final cooling of 60°C for 30 seconds. The 20 ul reaction contained 1X EvaGreen® SsoFast™ (Bio-Rad). The concentration of primer was listed in Table 1.
Melting Analysis
The melting curve was run after cycling by a ramping from 60ªc to 95ºC (with a temperature transfer speed of 0.02ºC/second).
RESULTS
Using the DNA of healthy controls, the amplification conditions and melting temperatures (Tm) of each segment of interest was determined (Table 1). Then, we combined different primers performing reactions with 2 (duplex) and 3 (triplex) sets of primers (Table 2), identifying the Tm of each product, in both multiplex qPCR (Figure 2 and 3).
Triplex qPCR products of control samples were analyzed on an 2% agarose gel, obtained distinguishable bands in each of the three reactions (Figure 4), allowing to correlate and confirm the results of dissociation temperature.
The sensitivity of the Triplex qPCR was evaluated by using DNA from a positive control and conducting the PCR with subsequent melting analisys on increasing four-fold dilutions. The mealting peaks could be accurately identified up to an initial DNA concentration of 2.47 ng / ml., after 35 cycles of amplification.
We attempt to develop a quadruplex qPCR, however it was not possible to display the 4 products with sufficient discrimination between each Tm.
The evaluation of the 35 DNA samples with the triplex qPCR established found 3 patients with partial microdeletions in AZFa region (Figure 5) and none microdeletions in other regions investigated (AZFb and AZFc), having an incidence of the AZFa microdeletion of 8.8% (Table 3).
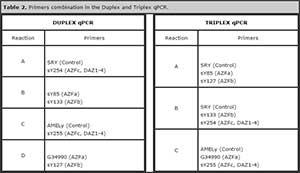
Table 2. Primers combination in the Duplex and Triplex qPCR.

Table 3. Oligozoospermic patients with AZFa microdeletion.
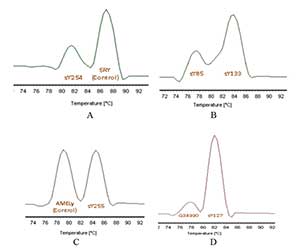
Figure 2. Melting Curves of the Duplex qPCR. The chart shows the melting temperatures of each set of primers when combined, also a deficency of the sY85, sY254 y G34990 primers is shown.
DISCUSSION
For development the multiplex real time PCR (qPCR), each set of primers were evaluated with DNA control of fertility men. The intercalating fluorescent agent –EvaGreen- was used to optimize the Tm of each PCR product.
With duplex qPCR, Tm of each product was identify, but with a reduced amplification efficiency in sY85 (AZFa), G34990 (AZFa) and sY254 (AZFc, DAZ1-4) sequences (Figure 3). The problem was solved by changing the concentration of the three sets of primers.
Using the optimized parameters in the duplex qPCR, we proceeded to test a triplex qPCR, combining primers as indicated in Table 1. Differentiable dissociation curves for each amplicon were obtained (Figure 4).
We try to establish a quadruplex qPCR, combining the primers as described by Kozina et al. (2011); however it was not possible to display the four products with sufficient discrimination between each temperature. This might give to the poor sensitivity of the thermocycler used.
Triplex qPCR sensitivity identifies accurate melting temperature with 2.47 ng/ml DNA initial concentration, after 35 amplification cycles. This sensitivity is lower than thareported by Guo et al. (2012) and Kozina et al. (2011); nevertheless, Guo et al. (2012) working with probes that give higher sensitivity and specificity than a DNA-binding dye such as EvaGreen.
The 35 oligozoospermic cases evaluated with optimized technique found three patients with partial AZFa microdeletions, corresponding to 8.8% incidence (Table 3). No case with microdeletions in AZFb and AZFc was found.
These results reveal a higher incidence than that reported in other studies such as Silber (2011), Kleiman et al. (2012), Behulova et al. (2010) and O’Flynn et al. (2010); nevertheless, like Fu et al. (2012), this study found only partial microdeletions and not total deletions. AZFa partial deletions prevalence have a wide range (0.2% -11%) (Kleiman et al. 2012), not defined by the few studies reported.
Studies report a AZFc microdeletions as the most frequent. The variation of the results found in this study could be a result of the geographic localization and racial characteristics of the studied population; however, to corroborate theses preliminary findings, it would be necessary to expand the number of samples.
The work regarding the incidence of AZF microdeletions have been mainly conducted on Asian and European populations. There are few studies published in Latin America as Lucena et al. (2004) and Fernández-Salgado et al. (2006), both reporting deletions in the AZFc region with a frequency of 6% and 3.4% respectively; but like this study, the number of cases included in both publications (97 and 29 respectively) was very low to determine the incidence.
Other genetic alterations involved in male infertility have been identified (Kozina et al., 2011; Buch et al., 2003; Kovac et al., 2013). This designed multiplex technique is restricted for identifying large deletions in the AZF section, like Kozina et al. (2011) and Simoni et al.(2001) protocols, it may be possible that the evaluated patients present other genetic problems in the Y chromosome that can not be identified with this technique.
This work would be the first Ecuadorian report about the optimization of a molecular technique to detect microdeletion in AZF region of Y chromosome.
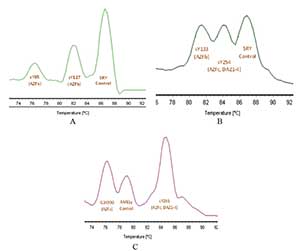
Figure 3. Melting Curves of the Triplex qPCR. The chart shows the differentiation of the melting curves of each set of primers when combined.
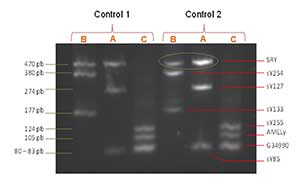
Figure 4. Observation of control samples amplicons in an electrophoresis gel. On the right side, the sequence names are describe and on the left, their weight is displayed.
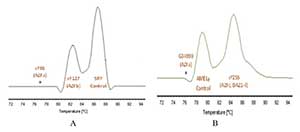
Figure 5. Microdeletions identified on the samples. Figure A and B show absence of sY85 and G34990 regions in the triplex qPCR.
CONCLUSIONS
The triplex qPCR optimizated allows the identification of microdeletions in AZFa, AZFb and AZFc region in infertile men and a better clinical management of the patient’s treatment decision. This report, done for the first in Ecuador, reveled a higher prevalence of microdeletion in the AZFa region in comparison with those previously described in other populations.
REFERENCES
Behulova R, Varga I, Strhakova L, Bozikova A, Gabrikova D, Boronova I, Repiska V. Incidence of Microdeletions in the AZF Region of the Y chromosome in Slovak Patients with Azoospermia. Biomed Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2011;155:33-8.
Medline Crossref
Buch B, Galán JJ, Lara M, Ruiz R, Segura C, Real LM, Martínez-Moya M, Ruiz A. Scanning of Y-chromosome azoospermia factor loci using real-time polymerase chain reaction and melting curve analysis. Fertil Steril. 2003; 80:907-13.
Medline Crossref
Elfateh F, Rulin D, Xin Y, Linlin L, Haibo Z, Liu RZ. Prevalence and patterns of Y chromosome microdeletion in infertile men with azoospermia and oligozoospermia in Northeast China. Iran J Reprod Med. 2014;12:383-8.
Medline
Fernández-Salgado E, Alvarez-Nava F, Borjas-Fajardo L, Osuna J, Gómez R, Zabala W, Zambrano M, Portillo-M MG. [Molecular analysis of microdeletions of the Y chromosome in Venezuelan males with idiopathic infertility]. Invest Clin. 2006; 47:395-403.
Medline
Fu L, Xiong DK, Ding XP, Li C, Zhang LY, Ding M, Nie SS, Quan Q. Genetic Screening for Chromosomal Abnormalities and Y chromosome Microdeletions in Chinese Infertile Men. J Assist Reprod Genet. 2012;29:521-7.
Medline Crossref
Guo Q, Lan F, Xu L, Jiang Y, Xiao L, Huang H, Zhou Y. Quadruplex real-time polymerase chain reaction assay for molecular diagnosis of Y-chromosomal microdeletions. Fertil Steril. 2012;97:864-9.
Medline Crossref
Khabour O, Fararjeh A, Alfaouri A. Genetic screening for AZF Y chromosome microdeletions in Jordanian azoospermic infertile men. Int J Mol Epidemiol Genet. 2014; 5:47-50.
Medline
Kleiman S, Yogev L, Lehavi O, Hauser R, Botchan A, Paz G, Yavetz H, Gamzu R.The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature (1994-2010). Fertil Steril. 2011;95:2005-12.
Medline Crossref
Kleiman S, Almog R, Yogev L, Hauser R, Lehavi O, Paz G, Yavetz H, Botchan A. Screening for partial AZFa microdeletion in the Y chromosome of infertile men: is it of clinical relevance? Fertil Steril. 2012;98:43-7.
Medline Crossref
Kovac J, Pastuszak A, Lamb D. 2013. The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil Steril. 2013;99:998-1007.
Medline Crossref
Kozina V, Cappallo-Obermann H, Gromoll J, Spiess AN. A One-Step Real-Time Multiplex PCR for Screening Y Chromosomal Microdeletions without Downstream Amplicon Size Analysis. Plos One. 2011; 6:e23174.
Medline Crossref
Krausz C, Hoefsloot L, Simoni M, Tüttelmann F, European Academy of Andrology, European Molecular Genetics Quality Network. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2:5–19.
Medline Crossref
Lucena E, Pérez C, Kent M. Determinación de Deleciones en el Cromosoma Y en Hombres Infértiles, Candidatos a Técnicas de Reproducción Asistida. NOVA. 2004; 2:16-27.
Link
Navarro-Costa P, Pereira L, Alves C, Gusmão L, Proença C, Marques-Vidal P, Rocha T, Correia SC, Jorge S, Neves A, Soares AP, Nunes J, Calhaz-Jorge C, Amorim A, Plancha CE, Gonçalves J. Characterizing partial AZFc Deletions of the Y Chromosome with amplicon-specific sequence markers. BMC Genomics. 2007;28:1-9.
Medline Crossref
O’Flynn K, Varghese A, Agarwal A. The genetic causes of male factor infertility: A review. Fertil Steril. 2010;93:1-12.
Medline Crossref
Rolf C, Gromoll J, Simoni M, Nieschlag E. Natural Transmission of Partial AZFb deletion of the Y chromosome over three generations: case report. Hum Reprod. 2002;17:2267-71.
Medline Crossref
Silber S. The Y chromosome in the era of the intracytoplasmic sperm injection: a personal review. Fertil Steril. 2011;95:2439-48.e1-5.
Medline Crossref
Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int J Androl. 2004;27:240-9.
Medline Crossref
Simoni M. Molecular diagnosis of Y chromosome microdeletions in Europe: state-of-the-art and quality control. Hum Reprod. 2001;16:402-9.
Medline Crossref
Sheikhha M, Zaimy MA, Soleimanian S, Kalantar SM, Rasti A, Golzade M, Hoseini Fahraji H. Multiplex PCR Screening of Y-chromosome microdeletions in azoospermic ICSI candidate men. Iran J Reprod Med. 2013;11:335-8.
Medline
Soares A, Costa P, Silva J, Sousa M, Barros A, Fernandes S. AZFb microdeletions and oligozoospermia - which mechanisms? Fertil Steril. 2012;97:858-63.
Medline Crossref
Suganthi R, Vijesh V, Jayachandran S, Fathima Benazir JA. Multiplex PCR based screening for microdeletions in azoospermia factor region of Y chromosome in azoospermic and severe oligozoospermic south Indian men. Iran J Reprod Med. 2013;11:219-26.
Medline
Suganthi R, Vijesh VV, Vandana N, Fathima Ali Benazir J. Y choromosomal microdeletion screening in the workup of male infertility and its current status in India. Int J Fertil Steril. 2014;7:253-66.
Medline
Yuen R, Merkoulovitch A, MacDonald JR, Vlasschaert M, Lo K, Grober E, Marshall CR, Jarvi KA, Kolomietz E, Scherer SW.. Development of a high-resolution Y-chromosome microarray for improved male infertility diagnosis. Fertil Steril. 2014;101:1079-85.e3.
Medline Crossref
Zaimy M, Kalantar SM, Sheikhha MH, Jahaninejad T, Pashaiefar H, Ghasemzadeh J, Zahraei M. The frequency of Yq microdeletion in azoospermic and oligospermic Iranian infertile men. Iran J Reprod Med. 2013;11:453-8.
Medline